Writing | Zhang Hui Editor | Zhou Yebin
Acute Lymphoblastic Lukemia (ALL) is the most common malignant tumor in children, with a peak incidence of 2 to 5 years old. When a child is diagnosed with ALL, parents can’t help but ask: “Why is my child so unfortunate”, or, “Doctor, how did my child get this disease”.
More and more evidence shows that: There is a significant genetic susceptibility to ALL in children compared with ALL in adults. Through Genome-Wide Association Studies (GWAS), 15 genetic variation loci associated with the pathogenesis of childhood ALL have been found so far, which are located in GATA3, ARID5B, IKZF1, CEBPE, PIP4K2A-BMI1, CDKN2A/2B, LHPP , ELK3, BAK1, IGF2BP1, USP7, IKZF3, ERG, TP63 and SP4, which increased the risk of childhood ALL to varying degrees.
However, these loci identified by GWAS are only indicative of increased risk. What mechanisms do these variants use to increase the risk, what kind of leukemia risk is associated with, and can they be prevented? These questions remain to be answered.
February 3, 2022, Professor Jun Yang from St. Jude Children’s Hospital and his team National Children’s Medical Center Shanghai Children’s Medical Center Zhang Hui< /strong>Doctor, and Professor Yue Feng from Northwestern University Feinberg School of Medicine and his team Yang Hongbo researcher, Luan Yu, Fudan University Obstetrics and Gynecology Hospital >Postdoctoral fellow, published an article entitled “Noncoding genetic variation in GATA3 increases acute lymphoblastic leukemia risk through local and global changes in chromatin conformation” in Nature Genetics.
The study found that the genetic variant rs3824662 in the GATA3 gene was associated with childhood Philadelphia chromosome-like acute lymphoblastic leukemia (Ph-like ALL) through deep targeted sequencing of ALL germline samples The risk of morbidity is highly correlated. Combined with omics experiments, it was found that the risk allele A of rs3824662 forms an enhancer at the locus, recruits the upstream transcription factor nuclear factor I/C, activates GATA3 transcription in cis, and then activates Ph-like characteristic genes by remodeling the spatial conformation of chromatin transcription, resulting in a Ph-like phenotype of B lymphocytes. Using a leukemia cell transformation model, as well as a zebrafish model, we found that GATA3 promotes the Ph-like ALL leukemia transformation process of B lymphocytes by activating CRLF2-JAK2-STAT5 signaling. This study provides a model reference for the systematic study of children’s genetic susceptibility to ALL, enriches the understanding of ALL prevention, and makes the identification of risk groups and early prevention more accurate.
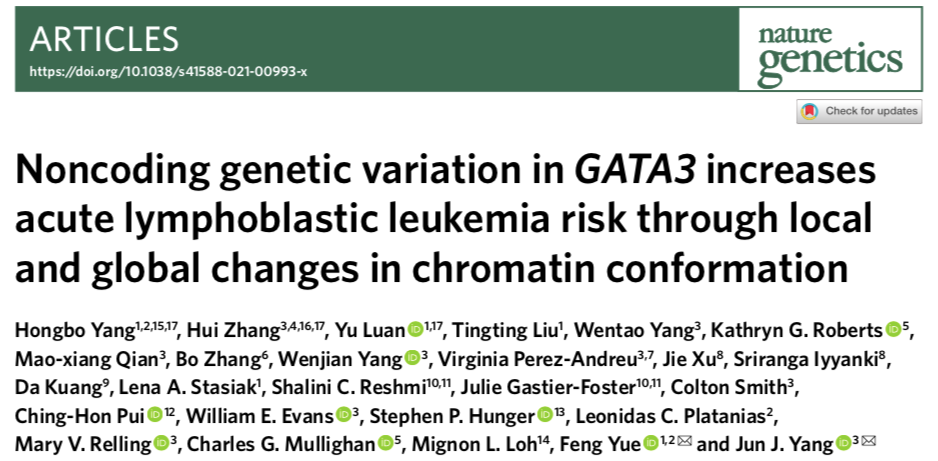
Childhood Philadelphia chromosome-like acute lymphoblastic leukemia (Ph-like ALL) is a group of ALL subtypes with similar expression profiles to Philadelphia chromosome-positive ALL, with poor prognosis. In order to clarify the genetic susceptibility to the pathogenesis of Ph-like ALL in children, the research team carried out deep targeted sequencing of the GATA3 locus of 5008 children with B-cell ALL (including 985 Ph-like ALL) on the basis of the previous study and susceptibility The results showed that the risk allele A of rs3824662 had the highest correlation, and the correlation was independent of gender, age, and race, suggesting the functional universality of this variant locus.
GATA3 is a T lymphocyte specific transcription factor. However, rs3824662 is not in the exon region of GATA3, so it does not affect GATA3 protein function. So the question arises, how does GATA3 rs3824662 in B-ALL affect the pathogenesis of Ph-like ALL in children? By integrating the Roadmap Epigenomics Project data and the luciferase reporter gene experiment, the research team found that the rs3824662 risk allele A forms an enhancer in B lymphocytes, which is a region characterized by chromatin opening and relaxation. More interestingly, in normal This phenomenon was found in both B lymphocyte strains and normal B lymphocytes from B-ALL patients.
rs3824662 is associated with Ph-like ALL susceptibility, and risk allele A confers enhancer activity and is associated with open chromatin state. A) Deep targeted sequencing revealed the GATA3 locus and the susceptibility to Ph-like ALL. B) Roadmap Epigenomics Project analysis shows that rs3824662 is located in a potential enhancer region of B lymphocytes. C) Validation of rs3824662 risk allele acquisition of enhancer activity in normal B lymphocytes GM12878. D-F) Epigenetic studies of normal B-lymphocyte lineages and normal B-lymphocytes from B-ALL patients show that the region where rs3824662 risk allele A is located is in an open chromatin state.
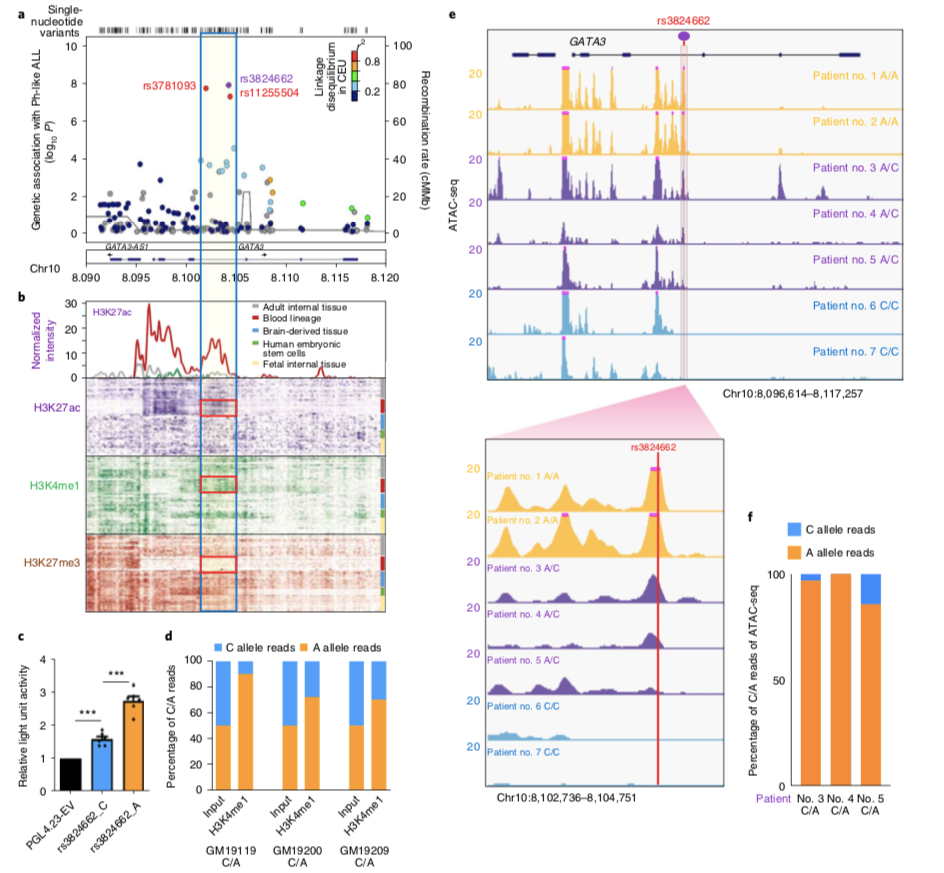
The enhancer activity of GATA3 rs3824662 risk allele A, which transcription factor is passed upstreamThe transcriptional activation function is generated, which genes are activated, and what characteristics are associated with Ph-like ALL generated by this transcriptional activation? To answer this question, the research team conducted a series of experiments. It was found by Capture-Seq and TADqPCR that rs3824662 activated GATA3 transcription in cis by interacting with the GATA3 promoter region, but had no effect on other genes in the same topologically related domain. Furthermore, through DNA binding prediction and ChIP-qPCR, overexpression and interference experiments, it was proved that the transcription factor NFIC binds to rs3824662 risk allele A but not allele C, and activates GATA3 transcription. The increased expression of GATA3 further binds to the regulatory regions of Ph-likeALL-related genes in B lymphocytes, especially the transcription initiation site, thereby making B lymphocytes susceptible to the Ph-likeALL phenotype.
rs3824662 risk allele A promotes the expression of genes characteristic of Ph-like ALL by cis-activating GATA3 transcription. A) The rs3824662 risk allele has an effect on GATA3 transcription only in the same topologically related domain. B) Chromatin conformation capture showing rs3824662 interacts with the GATA3 promoter. C) The luciferase reporter gene assay shows that the enhancer activity of rs3824662 risk allele A increases with co-expression of NFIC. D) Both NFIC interference or knockdown significantly reduced GATA3 expression. E-H) In B lymphocytes, GATA3 overexpression binds to genes characteristic of Ph-like ALL, especially at their transcription initiation sites.
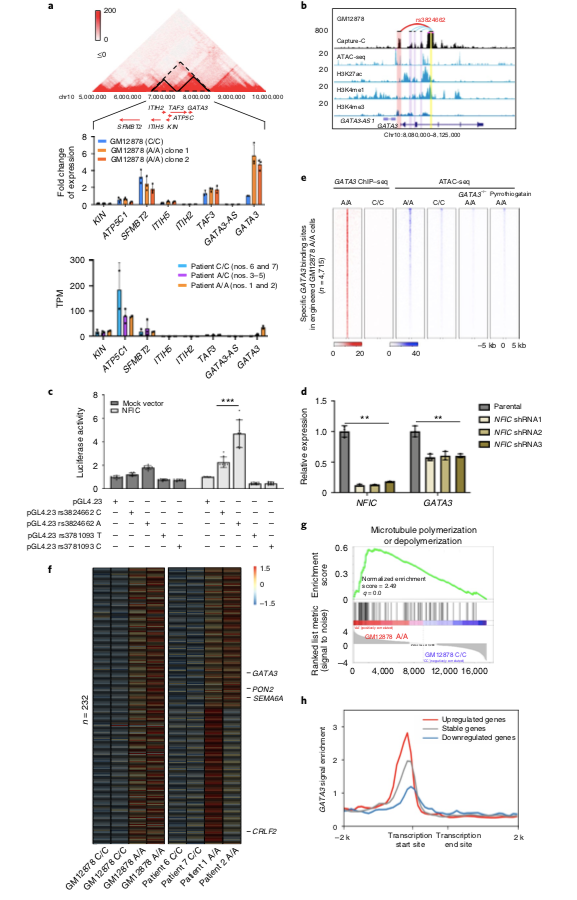
GATA3 rs3824662 risk allele A was only associated with GATA3 expression, but did not affect GATA3 protein function. Why does the GATA3 rs3824882 risk allele A produce such a dramatic response in B lymphocytes, and what are the secrets? The research team used the HiC and ChiPseq data of CRISPR/Cas9 gene-edited B lymphocytes and B-ALL cells to perform conformational analysis and found that the rs3824662 allele changed from C to A, and the chromatin spatial conformation of B lymphocytes was converted, especially in the Ph-like ALL-related genes.
GATA3 remodels the spatial conformation of chromatin in B lymphocytes. A-B) HiC of CRISPR/Cas9 gene-edited GM12878 (A/A) cells versus GM12878 (WT) cells revealed that the rs3824662 risk allele A remodeled B lymphocyte chromatin conformation. C-D) Chromatin conformational remodeling is particularly prominent on genes characteristic of Ph-like ALL, such as PON2.
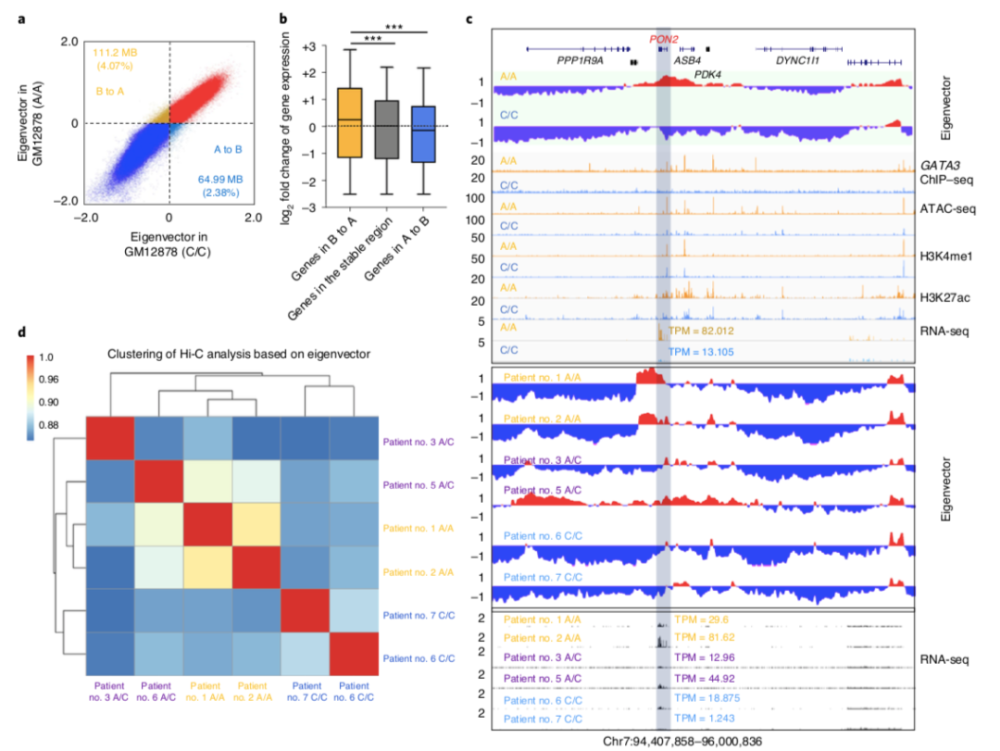
What kind of conformational change produces the characteristics of Ph-like ALL?
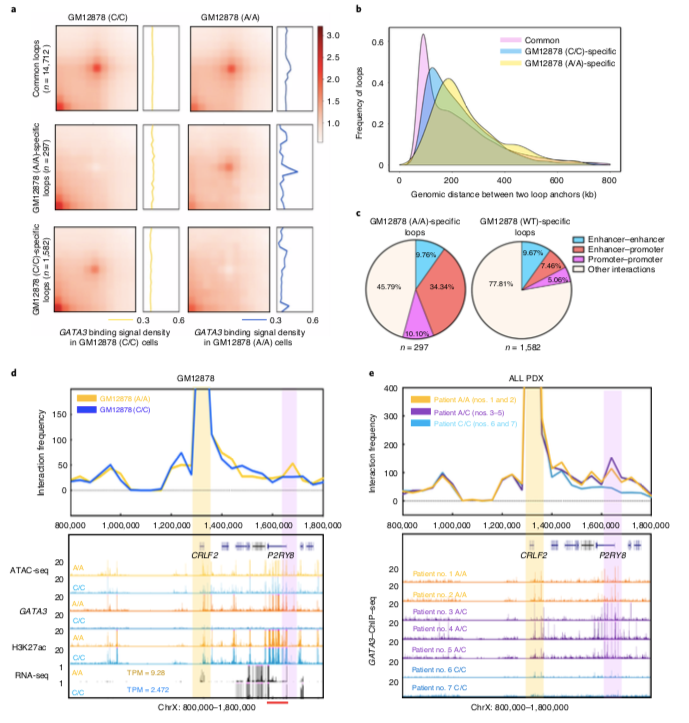
GATA3 increases enhancer-promoter interaction and promotes transcription and interaction of genes characteristic of Ph-like ALL. A-C) In CRISPR/Cas9 gene-edited B lymphocytes, HiC data show that the rs3824662 risk allele A can more significantly affect the interactions between gene regulatory domains, especially between enhancers and promoters. DE) In addition to its effect on the expression of genes characteristic of Ph-like ALL, the study also found that the carriage of the rs3824662 risk allele A is highly enriched near the partner gene of the fusion gene and forms a chromatin between the two partner genes loop, which promotes the occurrence of fusion genes.
Research on molecular mechanisms of genetic susceptibility hopes to benefit children with potential ALL at the root. In order to answer this question more deeply, based on the above results, the research team studied the GATA3-inducible expression cell line controlled by doxycycline and found that during the induction of GATA3, the expression of CRLF2 was up-regulated, and JAK2-STAT5 was activated at the same time. The leukemic transformation model further confirmed that GATA3 promoted the leukemic transformation of lymphocytes by activating the CRLF2-JAK2-STAT5 signal transduction pathway. This phenomenon has also been verified in the zebrafish experiment of GATA3.
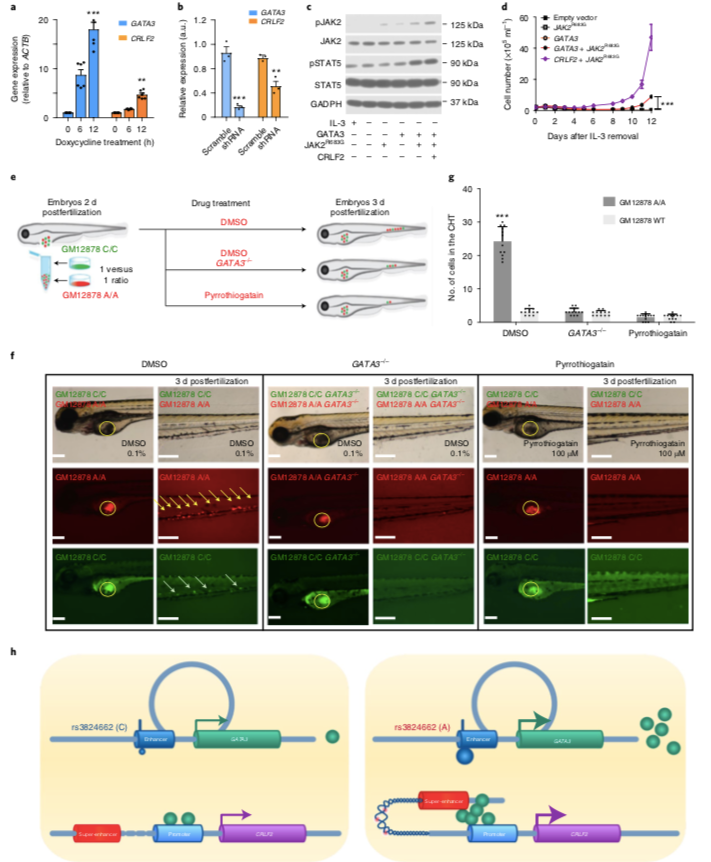
GATA3 promotes B lymphocytic leukemia transformation by activating the CRLF2-JAK-STAT signal transduction pathway. A-B) GATA3 directly regulates CRLF2 transcription. C-D) GATA3 promotes B lymphocytic leukemia transformation by activating the JAK2-STAT5 signaling pathway. E-G) GATA3-overexpressing cells have more advantages in hematopoietic proliferation. H) Schematic diagram of GATA3 rs3824662 risk allele susceptibility to B-ALL.
To sum up,this study focused on the susceptibility loci from the perspective of clinical problems, and clarified its susceptibility mechanism through basic research. abnormally high tableDa, by remodeling the spatial conformation of B lymphocytes chromatin, enriched and bound to the promoter/enhancer region of Ph-like characteristic genes, initiating its transcription, and then leading to leukemic transformation of B lymphocytes by activating the JAK2-STAT5 signaling pathway< /strong>.
What’s more interesting is that the research team’s research around this genetic variant also found: GATA3 rs3824662 genetic variant is highly correlated with poor early treatment response to childhood B-ALL< strong>, mechanism studies show that GATA3 activates JAK2-STAT3-autophagy signal transduction, making ALL cells resistant to asparaginase, resulting in poor early treatment response (J Natl Cancer Inst, 2021; Clin Transl Med , 2022).
This series of researches on the genetic variation of GATA3 rs3824662 really starts from the clinic, turns to scientific research, and then returns to the clinic to guide the clinical precision stratification and prediction, improve the clinical diagnosis and treatment of ALL, and provide systematic research on children Genetic susceptibility to ALL provides a model reference. Despite this, we still cannot answer the question of the patient’s parents very well. Only through delicate and meticulous research can we get closer to the truth, so as to better serve the clinic and improve the prevention and treatment of ALL in children.
Writing
Editor
Production
Typesetting | Car Jie proofreading | uu